KLIFS QuickStart
This example-driven tutorial will introduce you to the functionalities of the Kinase-Ligand Interaction Fingerprints and Structures database (KLIFS).
The topics are introduced in this tutorial:
You can use the menu below to go to a specific topic.
The topics are introduced in this tutorial:
- Searching for ligands by using SMILES
- Searching for ligands by drawing structures
- Structure filtering based on pocket composition and structure properties
- Ligands filtering based on interaction pattern and ligand binding mode
- Comparing ligands using the 3D Viewer
You can use the menu below to go to a specific topic.
1) Simple search
As the name suggests, KLIFS is a structural database that is focused on the interactions between protein kinases and the ligands that bind to them. KLIFS is designed in such a way that its users can quickly and easily search through all currently available protein kinase structures. This tutorial starts with finding all protein kinase structures with Imatinib (PDB: STI; marketed by Novartis as Gleevec). There are 3 ways to do that.

1.1.1) Type 'STI' or 'sti' in the PDB-code.
1.1.2) If you don't know the 3-letter HETATM PDB code for Imatinib, but do know a kinase structure in which Imatinib is co-crystallized, type its 4-letter PDB-code in the PDB-code box.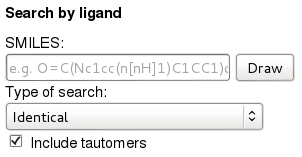
You can also search for a ligand by SMILES.
1.2.1) Go to the search page
1.2.2) Under 'Search by ligand' past the SMILES 'O=C(Nc1cc(Nc2nc(c3cnccc3)ccn2)c(cc1)C)c4ccc(cc4)CN5CCN(CC5)C' in the SMILES field. Along identical search, the database supports identity, (dis)similarity, and substructure searches. To view the supported search options, click on the 'Type of search' menu under the SMILES field.
1.2.3) Press 'Search!' at the bottom of the page.
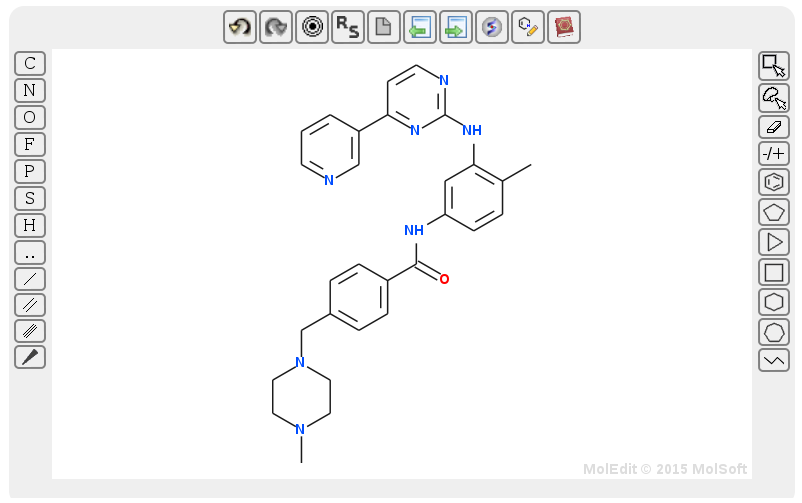
1.3.1) Go to the search page.
1.3.1) Press the draw button and draw the structure of Imatinib. Alternatively, you can click on the book icon (within the draw field) and type the name of the ligand to draw its structure. Just like SMILES search (step 1.2), structure search supports identity, (dis)similarity, and substructure searches.
1.3.1) Once the structure is drawn, press 'Search!'
Example of a structure with Imatinib:

To quickly view a ligand structure, move the mouse over the ligand name - in this case '4-(4-methyl-piperazin-1-ylmethyl)-n-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)-phenyl]-benzamide'. To download a structure-ligand complex in MOL2 format (as aligned and prepared by KLIFS), click on . You can also download all results as an Excel, CSV, zip file, or MOE database by clicking on one of the 3 download buttons at the bottom of the page. To view detailed structural data about a structure-ligand complex, click on . In order to add a particular structure-ligand complex to Favorites click on
. You can also download all results as an Excel, CSV, zip file, or MOE database by clicking on one of the 3 download buttons at the bottom of the page. To view detailed structural data about a structure-ligand complex, click on . In order to add a particular structure-ligand complex to Favorites click on  .
.
1.1) Using ligand PDB code and the PDB-code box at the upper right corner

1.1.1) Type 'STI' or 'sti' in the PDB-code.
1.1.2) If you don't know the 3-letter HETATM PDB code for Imatinib, but do know a kinase structure in which Imatinib is co-crystallized, type its 4-letter PDB-code in the PDB-code box.
1.2) Using the ligand structure in SMILES format

You can also search for a ligand by SMILES.
1.2.1) Go to the search page
1.2.2) Under 'Search by ligand' past the SMILES 'O=C(Nc1cc(Nc2nc(c3cnccc3)ccn2)c(cc1)C)c4ccc(cc4)CN5CCN(CC5)C' in the SMILES field. Along identical search, the database supports identity, (dis)similarity, and substructure searches. To view the supported search options, click on the 'Type of search' menu under the SMILES field.
1.2.3) Press 'Search!' at the bottom of the page.
1.3) By drawing the ligand structure or searching using the ligand name

1.3.1) Go to the search page.
1.3.1) Press the draw button and draw the structure of Imatinib. Alternatively, you can click on the book icon (within the draw field) and type the name of the ligand to draw its structure. Just like SMILES search (step 1.2), structure search supports identity, (dis)similarity, and substructure searches.
1.3.1) Once the structure is drawn, press 'Search!'
1.4) Results
After a search query is submitted, the results page will show a table with all protein kinase structures with Imatinib. On the top of the page the amount of entries and PDB structures is displayed. Example of a structure with Imatinib:

To quickly view a ligand structure, move the mouse over the ligand name - in this case '4-(4-methyl-piperazin-1-ylmethyl)-n-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)-phenyl]-benzamide'. To download a structure-ligand complex in MOL2 format (as aligned and prepared by KLIFS), click on
 . You can also download all results as an Excel, CSV, zip file, or MOE database by clicking on one of the 3 download buttons at the bottom of the page. To view detailed structural data about a structure-ligand complex, click on . In order to add a particular structure-ligand complex to Favorites click on
. You can also download all results as an Excel, CSV, zip file, or MOE database by clicking on one of the 3 download buttons at the bottom of the page. To view detailed structural data about a structure-ligand complex, click on . In order to add a particular structure-ligand complex to Favorites click on  .
.
2) Advanced search
KLIFS allows advance structure and ligand searches. For example, one could search for structures of a protein kinase family within certain resolution that contain ligands that make interactions with certain residues and bind in specific pockets. Note that you can combine step 1.2 Using SMILES and the SMILES field and step 1.3 Using structures and the draw field with the steps described here. Please refer to the FAQ for more information regarding the pocket definitions and residue numbering. Let's look at a couple of cases.
2.1.1) Go to the search page.
2.1.1) Under 'Search by ligand binding mode' check 'Front pocket' and 'Back pocket IV'.
2.1.2) Select 'Human' by Organism under 'Search by classification'.
2.1.2) Click on at 'Search by structure properties' and then click on .
at 'Search by structure properties' and then click on .
2.1.2) Click on 'Search!'.
2.2.1) Go to the search page.
2.2.2) Click on at 'Search by binding pocket composition (sequence)' and then select 'T' for GK at position 45 (GK.45) as shown below.
at 'Search by binding pocket composition (sequence)' and then select 'T' for GK at position 45 (GK.45) as shown below.

2.2.3) Click on at 'Search by ligand-kinase interaction pattern' and select '*' for GK.45, 'H' for hinge.46, 'X' for hinge.47 and 'H' for hinge.48.
at 'Search by ligand-kinase interaction pattern' and select '*' for GK.45, 'H' for hinge.46, 'X' for hinge.47 and 'H' for hinge.48.
2.2.4) Click on 'Search!'.
2.3.1) Go to the search page.
2.3.2) Click on I5 under "Search by pocket waters:".
2.3.3) Draw bosutinib or use the "Book icon" to search for the structure of bosutinib.
2.3.4) Select "MFP similarity (≥0.3)" under "Type of Search".
2.3.5) Click on 'Search!' to search for kinase structures with bosutinib or bosutinib analogues that have the conserved water at position I5.
2.1) Case 1
Suppose we are interested in ligands that bind to human protein kinase structures in at least one of the front pockets and back pocket IV and meet the Lipinski's rule of 5. 2.1.1) Go to the search page.
2.1.1) Under 'Search by ligand binding mode' check 'Front pocket' and 'Back pocket IV'.
2.1.2) Select 'Human' by Organism under 'Search by classification'.
2.1.2) Click on
2.1.2) Click on 'Search!'.
2.2) Case 2
Suppose we are interested in ligands that make H-bond interaction with 2 outer hinge residues and no interaction with the middle residue (hinge.47). Furthermore, we want the ligands to bind to Threonine (Thr) gatekeeper. Let's see if we can find such ligands.2.2.1) Go to the search page.
2.2.2) Click on

2.2.3) Click on
2.2.4) Click on 'Search!'.
2.3) Case 3
Bosutinib is known to have a selectivity patterns due to conserved waters in the binding site (see Levinson & Boxer). The most important water molecules is at position I5 (called W1 in Levinson & Boxer).2.3.1) Go to the search page.
2.3.2) Click on I5 under "Search by pocket waters:".
2.3.3) Draw bosutinib or use the "Book icon" to search for the structure of bosutinib.
2.3.4) Select "MFP similarity (≥0.3)" under "Type of Search".
2.3.5) Click on 'Search!' to search for kinase structures with bosutinib or bosutinib analogues that have the conserved water at position I5.
3) 3D Viewer
Suppose we want to compare the structures and the binding mode of 3 different ligands in Abl structures with DFG-in conformations. In KLIFS this can be easily achieved with the 3D viewer. Let's see how.
3.1) Go to the search page.
3.2) Under 'Search by classification' select 'Abl'.
3.3) Under 'Search by structural annotation' select 'IN' for the DFG conformation, and set 'Ligand-bound' to 'Yes'.
3.4) Click on 'Search!'.
All Abl structures with DFG-in conformations and ligands are now shown. We can now compare the binding modes of different ligands:
3.5) Click on at '2f4j' chain 'A'; '2g2i' chain 'A' and '4twp' chain 'A'. You should now see 3 yellow stars.
at '2f4j' chain 'A'; '2g2i' chain 'A' and '4twp' chain 'A'. You should now see 3 yellow stars.
3.6) Go to the favorites page to confirm that all selected structures are indeed in the list.
3.7) Click on at axitinib.
3.8) On the right, you will see a figure of the ligand in the binding pocket. Click on the 3D button ( ).
).
3.9) A new window will open containing the 3D viewer portraying the protein in cartoon (using the KLIFS coloring for the binding site) and the ligand displayed as sticks in the binding site.
3.10) Click on the star right of 'Favorites'.
3.11) Select show to compare the ligands. You can use the options below the viewer to adjust the view, colors, and representation or add the protein surface.
3.1) Go to the search page.
3.2) Under 'Search by classification' select 'Abl'.
3.3) Under 'Search by structural annotation' select 'IN' for the DFG conformation, and set 'Ligand-bound' to 'Yes'.
3.4) Click on 'Search!'.
All Abl structures with DFG-in conformations and ligands are now shown. We can now compare the binding modes of different ligands:
3.5) Click on
 at '2f4j' chain 'A'; '2g2i' chain 'A' and '4twp' chain 'A'. You should now see 3 yellow stars.
at '2f4j' chain 'A'; '2g2i' chain 'A' and '4twp' chain 'A'. You should now see 3 yellow stars.3.6) Go to the favorites page to confirm that all selected structures are indeed in the list.
3.7) Click on at axitinib.
3.8) On the right, you will see a figure of the ligand in the binding pocket. Click on the 3D button (
 ).
).3.9) A new window will open containing the 3D viewer portraying the protein in cartoon (using the KLIFS coloring for the binding site) and the ligand displayed as sticks in the binding site.
3.10) Click on the star right of 'Favorites'.
3.11) Select show to compare the ligands. You can use the options below the viewer to adjust the view, colors, and representation or add the protein surface.