- 01-May-2023KLIFS @ RSC symposium on kinase inhibitor design
- 05-Jan-20232023 - a sneak peek
- 02-Apr-2022KLIFS workshop @ May 19th
- 01-Feb-2022Alternative coloring scheme
- 23-Dec-2021Five "hidden" KLIFS features
- 08-Dec-2021MGMS Frank Blaney award!
- 17-Aug-2021Powerful analyses using KLIFS data in KNIME
- 09-Jul-2021Modified residues and many more clinical candidates
- 03-May-2021Granular control over the kinase conformation
- 22-Apr-2021Drugs and clinical candidates
- 15-Jan-2021KLIFS presentation @ February 10th
- 21-Oct-2020KLIFS v3 in Nucleic Acids Research
- 09-Sep-2020Release version 3.0 - the overhaul release
- 31-Oct-2019Trends in Pharmacological Sciences publication (open access)
- 04-Sep-2019Secure browsing with KLIFS
- 01-Aug-2018Release version 2.4 - the quality and stability release
- 30-May-2017Release version 2.3 - the atypical Protein Kinase (aPK) release
- 08-Feb-2017Publication and release 3D-e-Chem-VM
- 16-Nov-2016Release version 2.2 - the Virtual Reality release
- 28-Jul-2016Release version 2.1 - The KNIME nodes release
- 24-Oct-2015Nucleic Acids Research publication
- 01-Jul-2015Release version 2.0
- 13-Aug-2013Release version 1.0
News
Index
KLIFS @ RSC symposium on kinase inhibitor design
From 9-10 May, the 10th SCI/RSC symposium on kinase inhibitor design will be held in London (UK). At this symposium, Albert J. Kooistra will give a talk on KLIFS. During the talk, he will introduce KLIFS and outline ongoing + upcoming work. The talk is entitled:
KLIFS: capturing three decades of kinase inhibitor design
You can still register! For more information go to the symposium website.
2023 - a sneak peek
🎉 Happy new year! 🎉
Another year of exciting research in the field of kinases has started. KLIFS started 2023 by expanding the list of annotated drugs and clinical candidates with 25 new kinase inhibitors whose molecular structure was previously undisclosed. This brings the total number of kinase inhibitors in phase 1-4 clinical trials to a whopping total of 629 (+ 12 selected inhibitors with preclinical studies)!
However, this year you can expect more updates in KLIFS:
- New pages to explore kinase drugs and clinical candidates
- A complete overhaul of the KLIFS alignment based on thousands of new kinase structures and AlphaFold models
- Annotation of UniProt residue numbers for each kinase matched with the KLIFS numbering scheme (including an API function to programatically retrieve this information)
- Expanding the annotation of covalent kinase inhibitors
- And much more...
KLIFS workshop @ May 19th
UPDATE: The presentation is now available on YouTube, check the playlist in our channel.
As part of the workshop series from the RSC Chemical Information and Computer Applications Group, the KLIFS team is joining forces with the Volkamer Lab and will give a 2-hour hands-on workshop. The meeting will be held online and is free of cost and open for all to join. You can register for the meeting via EventBrite.For more information, see the abstract below.
May 19th 2022
- PST time @ 6am
- EST time @ 9am
- UK time @ 2pm
- CET time @ 3pm
- IST time @ 6:30pm
- CST time @ 9pm
KLIFS: making kinase structures work
Albert J. Kooistra, University of Copenhagen, Dominique Sydow* & Andrea Volkamer, Charité – Universitätsmedizin Berlin (*currently: Sosei Heptares)
Over the past three decades, six thousand structures of the catalytic kinase domain have been made publicly available via the Protein Data Bank. But to what extent are we making use of this wealth of information? In order to harness this data in a better way and to make it readily available for all to use in their research, KLIFS was constructed. KLIFS, i.e. the Kinase–Ligand Interaction Fingerprints and Structures database, is a structural kinase database that systematically collects and processes all structures of the catalytic kinase domain. With the database, you can - for example - easily get a complete overview of all structures, search for ligands with a specific binding mode, identify analogs or your ligands of interest, collect data for your data mining and machine learning applications.
For this workshop, the developers of KLIFS have teamed up with the Volkamer Lab and therefore the workshop will be divided into two segments. First, Albert J. Kooistra will give an introduction to KLIFS and demonstrate different functionalities of the KLIFS website and the integration of KLIFS in KNIME via the 3D-e-Chem nodes. In the second half, Andrea Volkamer and Dominique Sydow will demonstrate, based on their new kinase-focused TeachOpenCADD workflow, how to assess kinase similarity from different data perspectives. They will emphasize their Python package KiSSim – a KLIFS-based kinase structural similarity fingerprint, and OpenCADD-KLIFS – a Python module to facilitate the integration of KLIFS data into kinase research workflows.
Alternative coloring scheme
In our latest NAR publication, we concluded the article with a sneak peek of the new features to come. One of those features was the implementation of an alternative, colorblind-friendly coloring scheme to address issues such as raised in this article by Dr. Robert Roskoski Jr.
After testing several colorblind-friendly palettes such as those from Okabe, Wong, and IBM, we finally decided on a palette from Paul Tol. This new coloring scheme is available by clicking on the gear icon in the menubar. Changing the coloring scheme impacts the schematic depictions, segment coloring on several pages, and the 3D viewer. In addition, interactions are now depicted using different shapes.
UPDATE (Feb 16th) : The 2D images and PyMOL sessions using the new Tol coloring scheme have been generated now and are available in the structure details view.
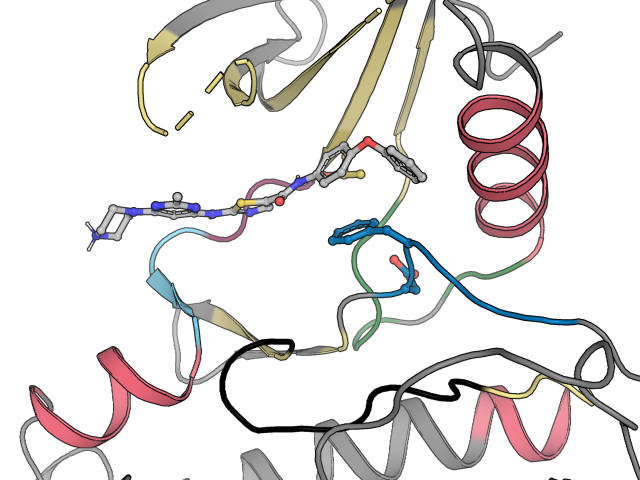
Five "hidden" KLIFS features
The KLIFS team wishes you happy and healthy holidays 🎄!
With this last post of 2021, we are sharing 5 tips and tricks to get more out of the KLIFS website:MGMS Frank Blaney award!
The Molecular Graphics and Modelling Society (MGMS) has presented Albert J. Kooistra with the 4th MGMS Frank Blaney award (formerly known as the MGMS Silver Jubilee Award) for his work on the research databases GPCRdb and KLIFS. This international prize has been awarded since 2007 to outstanding researchers with <10 postdoctoral years of experience in the field of molecular modelling and related areas (source MGMS). The accompanying monetary prize will go to continued hosting and maintenance of KLIFS.
For more information visit the MGMS website: https://www.mgms.org/WordPress/prizes/
Powerful analyses using KLIFS data in KNIME
KNIME is a powerful analytics platform and is freely available at https://knime.com. What not everyone knows, is that as part of the 3D-e-Chem project, we have developed KNIME nodes that directly interface with KLIFS. This enables everyone to create powerful (structural) kinase analysis workflows with minimal effort.
In this YouTube demonstration video, we show you how to add the KLIFS nodes to your KNIME setup. Subsequently, we demonstrate the functionality of these nodes by creating a simple, yet powerful, similarity search workflow.
3D-e-Chem publications on the developed KNIME nodes:
Modified residues and many more clinical candidates
Extensive annotation efforts have led to the addition of more than a hundred additional kinase inhibitors that either are or have been been in clinical trials. These kinase inhibitors have been complemented with bioactivity data from ChEMBL and, of course, structural data from KLIFS. This extra data is now available via the Drugs and Clinical candidates page and the API using the drug_list call.
Based on feedback from the KLIFS community, we have also added a new option to KLIFS that investigates if modified residues are present in a structure. These modification can for example be the phosphorylation or sulfation of residues in the structure. This information is now listed under the "Binding pocket sequence" segment of the structure detail page. See for example, this entry for PDB 2CJM - a CDK2 structure with phosphorylated residues.
This information is also available via a new API call: the structure_modified_residues call. The API call for PDB 2CJM (structure ID: 4406) would be: https://klifs.net/api_v2/structure_modified_residues?structure_ID=4406.
Granular control over the kinase conformation
Since the initial creation of the KLIFS classifiers for the DFG and αC-helix, the number of available kinase strutures has more than doubled. An update of these classifiers was therefore essential. After iterative rounds of data curation, data generation, and data mining, robust new models were obtained that tremendously improve the classification of the kinase conformation in KLIFS!
In addition, a new set of properties are now calculated for each kinase structure. These variables correspond to the positioning of the activation loop and the αC-helix, formation of the salt bridge between the conserved lysine (17) and glutamate (24), and the rotation of the aspartate and phenylalanine of the DFG. These properties can now be searched via the new "conformation" section on the search page and allow for granular control over these parameters (see screenshot below).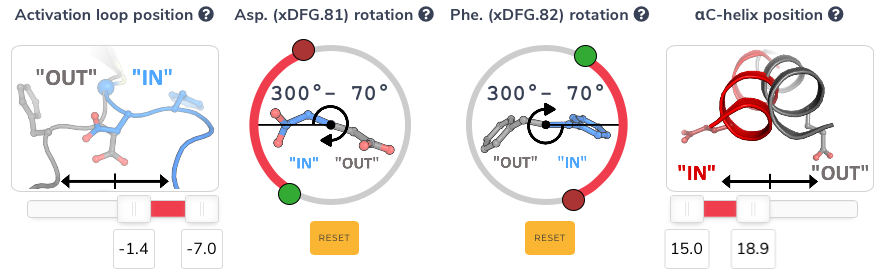
Finally, the API is being updated and a beta version is now available via the menu system. New calls have been added for the drugs and clinical inhibitors and the new conformational annotations. The latter includes a few bonus properties, such as the Möbitz dihedral and the rotational angle of the activation loop.
Please let us know what you think (Contact us! option in the menu under More)!
PS Big thanks to the Volkamer Lab for beta testing these new features!
Drugs and clinical candidates
The small molecule drugs and clinical candidates (available via "Drugs" in the menu) have been updated.
In addition to the latest data from the PKIDB with additional KLIFS curation and structural annotations, also new candidates whose structures were disclosed at AACR21 (such as Merck's M4205 and Pfizer's PF-07104091) have been added. And to provide an even more complete overview, all kinase inhibitors with an approved INN name that were not yet present have also been added.
KLIFS presentation @ February 10th
UPDATE: The presentation is now available on YouTube, check the playlist in our channel.
At the next meeting of the Cambridge Cheminformatics Network, Albert J. Kooistra is one of three invited presenters. He will give an overview of KLIFS, cover the latest updates, and (if time allows) give a short demonstration.
The meeting will be held online (Zoom) and is free of cost and open for all to join. You can register for the meeting via http://www.c-inf.net.
10 February 2021
- PST time @ 8am
- EST time @ 11am
- UK time @ 4pm
- CET time @ 5pm
- IST time @ 9:30pm
- CST time @ 0am (+1)
KLIFS v3 in Nucleic Acids Research
The release of version 3.0 of KLIFS is accompanied by a publication in Nucleic Acids Research (database edition 2021). The article has just been published online and is available to all (open access) at Nucleic Acids Research!
In the article we look back at how the research community (i.e you) has been using KLIFS, describe how KLIFS has changed throughout the years and what new features have been added. We conclude the article with a few case studies with which we demonstrate some of the new functionalities of KLIFS.
Thanks to all of you for using KLIFS and for your vital feedback!
DOI link to the article: doi.org/10.1093/nar/gkaa895
Release version 3.0 - the overhaul release
A major KLIFS release with a new website, a new domain (klifs.net), a new logo, and new features. You can still find all the functionalities from the previous website in this new design, but a lot of new things have been added. One of the major additions is the drugs page based on data from the PKIDB that allows you to quickly go from a drug to all related information in KLIFS.
In a few weeks, we will update you with more information on this release. In the meantime, please test the site and provide us with feedback (via the Contact us! option in the menu).
Trends in Pharmacological Sciences publication (open access)
The enrichment of KLIFS with the structures of atypical kinases in 2017 enabled us to finally compare atypical and eukaryotic protein kinases in a structure-centered manner. For this publication we have investigated their sequences, hydrophobic spines, mutation and SNP hotspots, and inhibitor interaction patterns. This work will hopefully contribute to the design of network medicines: targeting multiple kinases across the barriers of these kinase classes!
Thanks to support from Amsterdam Data Science we could make this work open access. Please contact us if you have any questions or comments!
The article is available here: Trends in Pharmacological Sciences
Secure browsing with KLIFS
It took a while, but our KLIFS server finally has its own SSL certificate. This means that your browser will now mark the connection to KLIFS as secure and will use encryption to communicate with our servers.
Please let us know if you encounter any issues (or have any suggestions) by filling out the contact form.
Release version 2.4 - the quality and stability release
This new release primarily contains new manually curated KLIFS content, updates data links and bugfixes. But that's not all, in order to improve the interactive exploration of the kinase-ligand structures we have included the NGL viewer on the kinase structure detail page in addition to the static images of the structure.As per usual, let us know if you spot any mistakes/issues or have any suggestions.
Added
- Interactive NGL viewer on the kinase structure detail page in addition to the static images
- Added last remaining eukaryotic and atypical protein kinases to KLIFS
- Columns with HET-codes of orthosteric and allosteric ligands to Excel/CSV download of search results
Changed
- Adjusted quality score for atypical kinases
- Optimized atypical reference alignment
- Updated ChEMBL to version v24.1
Fixed
- Removed deprecated structures from statistics overview
- An issue with pasting SMILES and then drawing the structure on the search page
- A bug that prevented the download of more than 7500 monomers simultaneously
- An issue that could erroneously adjust atom coordinates during ligand processing
- The misclassification of some kinase structures
- Incorrect DFG conformation classification for some structures
- Incorrect interpretation of PDB codes as numbers in scientific notation for CSV/Excel download
Release version 2.3 - the atypical Protein Kinase (aPK) release
With this new release also aPK structures (currently 181 PDBs) have been added to KLIFS after thoroughly refining their reference alignments. All aPK families with a catalytic kinase domain and ePK-like fold (ABC1, PIKK, PI(3)K, and RIO) are now part of KLIFS. The other atypical families (Alpha, Brd, PDHK, and TIF1) are omitted due to the absence of catalytic kinase domains or because they have a non-ePK fold. Please let us know if you spot any mistakes/issues or have any suggestions.
Added
- Atypical protein kinases have been added
- ChEMBL (p)EC50 bioactivity data points
- Search by "Release date" (under structure properties)
- Search now contains the option to include deprecated/superseded structures
- Downloads now contain the 3-letter PDB-codes for the ligands
Changed
- Updated ChEMBL to version v23
- Fully updated news overview
- Direct links instead of buttons to detail pages for optimal page indexing
- Alphanumerical sorting of kinase names, families, and groups
- Improved the download feature - performance
- Updated KLIFS knime nodes
Fixed
- Bug that made the close button of the search results kinome float away on wide screens.
- Too stringent mobile landing page redirect
- Erroneous ligand annotations
- Manually adjusted one structure due to different chain annotations
- Added a few missing human kinase structures
- The web services did not return "apo" structures
- Segmentation issues with partial alternate models and interaction fingerprints
- Added missing "master" SMILES for a few ligands
- Bug on the search page for IE11
- Bug resulting in all potential interactions for three structures
Publication and release 3D-e-Chem-VM
The 3D-e-Chem team has published its virtual cheminformatics virtual machine in the ACS Journal of Chemical Information and Modeling!This publication also describes the release of the KNIME nodes for the KLIFS database including example workflows using the KLIFS nodes.
Abstract:
3D-e-Chem-VM is an open source, freely available Virtual Machine (https://3d-e-chem.github.io/3D-e-Chem-VM/) that integrates cheminformatics and bioinformatics tools for the analysis of protein-ligand interaction data. 3D-e-Chem-VM consists of software libraries, and database and workflow tools that can analyze and combine small molecule and protein structural information in a graphical programming environment. New chemical and biological data analytics tools and workflows have been developed for the efficient exploitation of structural and pharmacological protein-ligand interaction data from proteome-wide databases (e.g. ChEMBLdb and PDB), as well as customized information systems focused on e.g. G Protein-Coupled Receptors (GPCRdb) and protein kinases (KLIFS). The integrated structural cheminformatics research infrastructure compiled in the 3D-e-Chem-VM enables the design of new approaches in virtual ligand screening (Chemdb4VS), ligand-based metabolism prediction (SyGMa), and structure-based protein binding site comparison and bioisosteric replacement for ligand design (KRIPOdb).
The article: http://dx.doi.org/10.1021/acs.jcim.6b00686
The virtual machine: https://3d-e-chem.github.io/3D-e-Chem-VM/
The KLIFS knime nodes: https://github.com/3D-e-Chem/knime-klifs/
Release version 2.2 - the Virtual Reality release
Added
- Virtual Reality: Google Cardboard viewer for mobile devices (visit KLIFS on your mobile phone). View the demo on YouTube
- A mobile landing page for easy access to the Virtual Reality viewer
- Weekly updates of PubMed identifiers
- Weekly updates article DOIs for PDB entries
- Weekly updated annotation of obsoleted and superseded PDB structures
- A new release of the KNIME nodes for KLIFS developed within the 3D-e-Chem project. Use KNIME to delve into KLIFS and extract all data and structures from within your own workflows! Just add our 3D-e-Chem repository to KNIME, install the KLIFS nodes, and you are good to go (for more information see the GitHub repository).
Changed
- 2D ligand depictions are now generated with CACTVS
- 2D depictions of allosteric ligands are now custom generated (replaces the depiction from the RCSB)
Fixed
- Small bug that caused an incorrect pocket overview for two structures
- Several missing images of the 2D molecular structure of the ligand
- Manually added two missing PDBs that were somehow not returned by the RCSB web services
- Small bug in the RCSB web services that caused an error upon species selection
- The interaction fingerprint was padded with too many zeroes in the MDB download option
- Bug that could lead to the faulty annotation of ligands with a PDB-code starting with a Q
- Issue when processing Ruthenium-containing ligands (again) that lead to the removal of the ligand
Acknowledgements
We would like to especially thank all KLIFS users that reported issues and/or requested new features. Your input is crucial to maintain a high quality database.Release version 2.1 - The KNIME nodes release
Added
- KNIME nodes for KLIFS developed within the 3D-e-Chem project. Use KNIME to delve into KLIFS and extract all data and structures from within your own workflows! Just add our 3D-e-Chem repository (https://3d-e-chem.github.io/updates) to KNIME, install the KLIFS nodes, and you are good to go (for more information see the GitHub repository).
- Web services including Swagger API definition (here)
- Kinome-through-time animation (here)
- Links to IUPHAR - Guide to pharmacology
- Predefined "lead-like properties" for the ligand properties search
- Included MGI nomenclature for the mouse kinases
- 3D viewer labels now shows KLIFS numbering
- Detailed interactions overview now includes original X-ray numbering
Changed
- Updated kinase classification and nomenclature
- Improved 2D depiction of ligands, however, we are still working on further improvement
- Updated the HGNC nomenclature and UniProt numbering
- Updated ChEMBL integration to version 21
- Included (p)Kd values in the bioactivity data overview
Fixed
- MOE database downloads did not always include all selected entries when NMR structures were encountered
- CSV downloads could erroneously mark all entries as "human"
- Issue for the 3D viewer on iOS devices: some structures were not visible
- Issue for the 3D viewer when the N-term of a structure was in a helix
Nucleic Acids Research publication
Nucleic Acids Research has accepted our manuscript regarding the new KLIFS database and website for publication in the database 2016 issue. You can freely access the article here: doi: 10.1093/nar/gkv1082.
Release version 2.0
The second version of KLIFS as described in the Nucleic Acids Research paper (doi: 10.1093/nar/gkv1082). KLIFS is now accessible at http://klifs.vu-compmedchem.nl and has many added features.
Added
- User-friendly and feature-rich web interface
- Weekly updates in sync with PDB
- Mouse eukaryotic kinases
- Automated analysis of DFG, αC-helix, and G-rich loop conformations
- Automated subpocket annotation of ligand binding modes
- Water cluster analysis
- Integrated webGL 3D viewer
- Improved atom and bond-typing (MOL2)
- ... (see the publication for more details)
Release version 1.0
The initial version of KLIFS as used for the ACS Journal of Medicinal Chemistry publication (doi: 10.1021/jm400378w). It contains the analysis of 1734 PDB structures covering 190 different human kinases.
KLIFS v1.0 is available as a dataset at Zenodo: